The TriVersa NanoMate® LESA® is the latest in chip-based electrospray ionization technology from Advion Interchim Scientific®. It combines the benefits of liquid chromatography, mass spectrometry, chip-based infusion, fraction collection and direct surface analysis into one integrated ion source platform. It allows scientists to obtain more information from complex samples than LC/MS alone.
Shotgun Lipidomics Analysis
Infusion based lipidomics analysis (shotgun Lipidomics) is an analysis strategy that involves the liquid extraction of lipids from samples such as blood, tissue homogenates or dried blood spots. This approach is suitable for a wide range of biomedical questions of lipid differentiation and lipid flux in the field of metabolic disease, neurological disorders, cancers or eye diseases. See how utilizing the Advion Interchim Scientific TriVersa NanoMate® can be used for high throughput shotgun Lipidomics analysis.
Lipid Species from Brain Tissue Sections Using LESA PLUS Liquid Extraction Surface Analysis PLUS LC Separation
Spatial lipid composition, distribution and regulation are very important factors for mediating lipid functionality and, when disrupted, can cause pathophysiological processes leading to cancer, obesity, atherosclerosis, and neurodegeneration. The novel LESAPLUS surface analysis approach combines the standard liquid extraction surface analysis with an additional step of a nano liquid chromatography separation. This combination is ideally suited to investigate small molecule drugs, metabolites or lipids from thin tissue sections.
Study of ionization efficiency on silicon-based, micro-fabricated electrospray nozzles for micro flow LC/ESI-MS applications
One of the benefits of nanoelecrtospray ionization (nanoESI) is the reported optimal ionization efficiency (equalized response) of chemically different analyses despite the presence of matrix components. However, outside of a very low flow rate of ca. 1-20 nL/min, the ionization efficiency drops exponentially and coupling higher flow rates from a typical liquid chromatography (LC) system to ESI loses most of the benefits such as enhanced sensitivity and reduced ion suppression in mass spectrometry analysis (LC/ESI/MS). The Advion silicon-based ESI Chip accommodates micro flow LC/MS applications at ionization efficiencies near optimal levels and comparable to nanoelectrospray ionization (nanoESI).
CHIPSOFT 10.0 WITH DEVELOPERS KIT: Customized method development for the TriVersa Nanomate
ChipSoftX is an entirely new operating software for the TriVersa NanoMate automated nanoelectrospray source. Besides improvement in program compatibility with Windows and integration of existing software features, it also provides access to the new Developers Kit – a platform for customized method development with direct access to robot controls allowing entirely novel analysis workflows such as LESAPLUS.
Screening and classifying small-molecule inhibitors of amyloid formation using ion mobility spectrometry–mass spectrometry
Young, LM, Saunders, JC, Mahood, RA, Revill, CH, Foster, RJ, Tu, LH, Raleigh, DP, Radford, SE, Ashcroft, AE
Nature Chemistry7,73–81doi:10.1038/nchem.2129
The search for therapeutic agents that bind specifically to precursor protein conformations and inhibit amyloid assembly is an important challenge. Identifying such inhibitors is difficult because many protein precursors of aggregation are partially folded or intrinsically disordered, which rules out structure-based design. Furthermore, inhibitors can act by a variety of mechanisms, including specific or nonspecific binding, as well as colloidal inhibition. Here we report a high-throughput method based on ion mobility spectrometry–mass spectrometry (IMS–MS) that is capable of rapidly detecting small molecules that bind to amyloid precursors, identifying the interacting protein species and defining the mode of inhibition. Using this method we have classified a variety of small molecules that are potential inhibitors of human islet amyloid polypeptide (hIAPP) aggregation or amyloid-beta 1-40 aggregation as specific, nonspecific, colloidal or non-interacting. We also demonstrate the ability of IMS–MS to screen for inhibitory small molecules in a 96-well plate format and use this to discover a new inhibitor of hIAPP amyloid assembly.
University of Southern Denmark, Ejsing Lab
What is the focus of your lab’s research?
Development and applications of lipidomic technology that allow global analysis of cellular lipidomes with the ultimate goal of improving the understanding of the regulation of lipid metabolism.
Why did you incorporate the TriVersa NanoMate?
I used the TriVersa NanoMate in Andrej Schevchenko’s laboratory at Max Planck Institute in Dresden. I learned that the instrument was far easier to use than nanoelectrospray needles. Additionally, the TriVersa NanoMate o ffers high throughput and is easy to teach others, especially students, how to use. You can rely on the TriVersa NanoMate to get the job done.
Who would you recommend to purchase the TriVersa NanoMate?
The TriVersa NanoMate is essential for any laboratory interested in lipidomics research for its ease of use, reliability and stable ion spray.
Do you have any publications or presentations using the TriVersa NanoMate?
Publication highlight
Lipid Molecular Timeline Profiling Reveals Diurnal Crosstalk between the Liver and Circulation
Sprenger et al. Cell Rep. 2021 34(5):108710
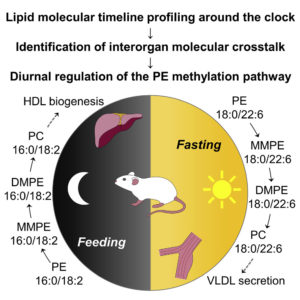
In-depth lipidomics and time-series analysis of fasting-feeding cycles and circadian rhythms provides insights into metabolic crosstalk between tissues.
Other Publications:
- Höring et al. Accurate quantification of lipid species affected by isobaric overlap in Fourier-transform mass spectrometry. Journal of Lipid Research. DOI: 10.1016/j.jlr.2021.100050
- Höring et al. Quantification of cholesterol and cholesterol ester by direct flow injection high-resolution fourier transform mass spectrometry utilizing species-specific response factors. Analytical Chemistry. DOI: 10.1021/acs.analchem.8b05013
- Artibani et al. Adipocyte-like signature in ovarian cancer minimal residual disease identifies metabolic vulnerabilities of tumor initiating cells. JCI Insight. DOI: 10.1172/jci.insight.147929
- Freyer et al. MIGA2 links mitochondria, the ER, and lipid droplets and promotes de novo lipogenesis in adipocytes. Mol Cell. DOI: 10.1016/j.molcel.2019.09.011
- Martinez-Montanes et al. Phosphoproteomic Analysis across the Yeast Life Cycle Reveals Control of Faty Acyl Chain Length by Phosphorylation of the Fatty Acid Synthase Complex. Cell Reports. DOI: 10.1016/j.celrep.2020.108024
University of Birmingham, Cooper Mass Spectrometry Group
What is the focus of your lab’s research?
Our research focuses on in situ analysis of intact proteins from biological substrates. We combine ambient surface techniques and ion mobility spectrometry with high resolution mass spectrometry.
We are particularly interested in native ambient mass spectrometry, in which folded proteins, protein assemblies and protein complexes are sampled directly from thin tissue sections. Native ambient mass spectrometry, such as liquid extraction surface analysis (LESA), is integrated with mass spectrometry imaging to provide simultaneous spatial and structural information.
We also apply LESA for the analysis of intact but unfolded proteins from a range of substrates including living microbial colonies growing on agar and other solid substrates, dried blood spots and tissue sections. The combination of LESA, ion mobility spectrometry and mass spectrometry enables the detection of hundreds of proteins.
Why did you incorporate the TriVersa NanoMate® into your laboratory
Initially, we purchased the TriVersa NanoMate® for direct infusion and coupling to LC and we still use the equipment for that purpose. Advion Interchim Scientific’s chip technology has revolutionized nanospray as far as ease of use. The ESI Chip™ is robust and bypasses any problems with non-uniformity. It allows us to move simply to the next nozzle if there is an issue with spray. The spray sensing capability is very clever and necessary for our overnight runs. More recently, we have used the LESA capability of the TriVersa NanoMate® for our in situ analyses of proteins in tissue, dried blood spots and microbial colonies.
To whom would you recommend the TriVersa NanoMate® for their research?
I would recommend the TriVersa NanoMate® to anyone with a mass spectrometer who uses nanoelectrospray.
Do you have any publications or presentations using the TriVersa NanoMate®?
Publication Highlight
Liquid Extraction Surface Analysis Mass Spectrometry of ESKAPE Pathogens
Havlikova et al. J Am Soc Mass Spec. 2021
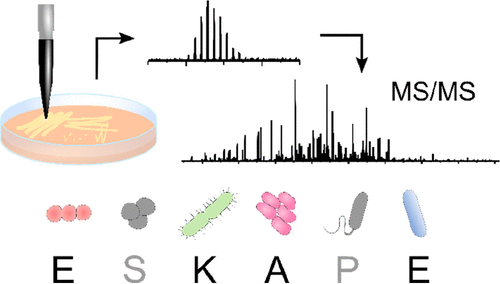
Top-down LESA MS/MS was used for protein identification in four ESKAPE pathogens as well as E. faecalis V583 and a clinical isolate of A. baumannii.
Other Publications:
- Hale et al. Native mass spectrometry imaging and in situ top-down identification of intact proteins directly from tissue. J Am Soc Mass Spec. DOI: 10.1021/jasms.0c00226
- Havlikova et al. Direct identification of bacterial and human proteins from infected wounds in living 3D skin models. Sci Rep. DOI: 10.1038/s41598-020-68233-6
- Haque et al. Self-incompatibility triggers irreversible oxidative modification of proteins in incompatible pollen. Plant Physiology. DOI: 10.1104/pp.20.00066
- Sisley et al. LESA cyclic ion mobility mass spectrometry of intact proteins from thin tissue sections. Anal. Chem. DOI: 10.1021/acs.analchem.9b05169
- Hale et al. Native LESA TWIMS-MSI: Spatial, conformational, and mass analysis of proteins and protein complexes. J Am Soc Mass Spec. DOI: 10.1021/jasms.9b00122
- Kocurek et al. Electroporation and mass spectrometry: A new paradigm for in situ analysis of intact proteins from living yeast colonies. Analytical Chemistry/ DOI: 10.1021/acs.analchem.9b04365
- Griffiths et al. Comprehensive LESA mass spectrometry imaging of intact proteins by integration of cylindrical FAIMS. Analytical Chemistry. DOI: 10.1021/acs.analchem.9b05124
- Havlikova et al. Quantitative imaging of proteins in tissue by stable isotope labeled mimetic liquid extraction surface analysis mass spectrometry. Analytical Chemistry. DOI: 10.1021/acs.analchem.9b04148
- Griffiths et al. LESA MS imaging of heat-preserved and frozen tissue: Benefit of multistep static FAIMS. Analytical Chemistry. DOI: 10.1021/acs.analchem.8b02739
- Rosting et al. High field asymmetric waveform ion mobility spectrometry in nontargeted bottom-up proteomics of dried blood spots. J Proteom Res. DOI: 10.1021/acs.jproteome.7b00746
- Sarsby et al. Liquid extraction surface analysis mass spectrometry coupled with field asymmetric waveform ion mobility spectrometry for analysis of intact proteins from biological substrates. Analytical Chemistry. DOI: 10.1021/acs.analchem.5b01151
- Griffiths et al. Liquid extraction surface analysis field asymmetric waveform ion mobility spectrometry mass spectrometry for the analysis of dried blood spots. Analyst. DOI: 10.1039/C5AN00933B
- Sarsby et al. Top-down and bottom-up identification of proteins by liquid extraction surface analysis mass spectrometry of healthy and diseased human liver tissue. J Am Soc Mass Spec. DOI:10.1007/s13361-014-0967-z
- Randall et al. Direct analysis of intact proteins from Escherichia coli colonies by liquid extraction surface analysis mass spectrometry. Analytical Chemistry. DOI: 10.1021/ac503349d
- Edwards et al. Compound heterozygotes and beta‐thalassemia: Top‐down mass spectrometry for detection of hemoglobinopathies. PROTEOMICS. DOI: 10.1002/pmic.201300316
- Martin et al. Dried blood spot proteomics: Surface extraction of endogenous proteins coupled with automated sample preparation and mass spectrometry. J Am Soc Mass Spec. DOI: 10.1007/s13361-013-0658-1
- Edwards et al. Hemoglobin variant analysis via direct surface sampling of dried blood spots coupled with high-resolution mass spectrometry. Analytical Chemistry. DOI: 10.1021/ac1030804
University of Bristol, School of Chemistry
Q: What is the focus of your lab’s research?
A: From drug-sized molecules to fragments, we analyze a little bit of everything. My primary research focuses around discovering the structure of proteins including enzyme structure and function in the development of antibiotics.
In addition, we have a collaboration with UCB, a global pharma focused on central nervous system and immunology disorders, involving library screening.
Q: Why did you incorporate the TriVersa NanoMate® into your laboratory?
A: We have a NanoMate 100 and a TriVersa NanoMate®. It is reasonably well-known that you need nanoESI for noncovalent interactions – ligand screening, but it was essential to my research to speed up and to automate the process. I had watched too many demonstrations that involved pulled capillaries, and I knew it would drive us crazy. A second instrument was absolutely vital to meet the needs of our collaboration with UCB.
Q: What benefits have you experienced with the TriVersa NanoMate®?
A: The main reason for purchasing the TriVersa NanoMate® is its automated nanoESI capability. The added benefit is its ease of use. We act as a service lab to the chemistry department, and often we have inexperienced users with a variety of different samples to be analyzed. The TriVersa NanoMate® makes it easy for users to do what needs to be done, very quickly.
Q: To whom would you recommend the TriVersa NanoMate® for their research?
A: I would recommend the TriVersa NanoMate® to anyone who is looking for an easy to use nanoelectrospray source, and who would like to automate and to speed up their process.
Institute for Research in Biomedicine (IRB Barcelona), Spain
What is the focus of your lab’s research?
Our goal is to provide the research community at IRB Barcelona and their co-workers with state-of-the-art tools and methodologies for the MS analysis of a broad range of biological species, from large proteins and DNA to small molecules. The final purpose is to get insight into these molecules’ identity, structure, interaction with other molecules and biological function in order to help in drug design, protein mechanism elucidation and in the search for biomarkers. We have implemented methods specialized in top-down proteomics and we are pioneers in this MS strategy in Spain.
As a core facility, we are responsible for working with different biologic molecules, and we are required to change methods constantly and efficiently.
How does the TriVersa NanoMate® align with your research goals?
Originally, we purchased the TriVersa NanoMate® for its chip-based direct infusion mode for noncovalent interaction analysis, but we learned quickly that it could be applied to other areas of our research. Prior to using the TriVersa NanoMate®, the steps involved in collecting fractions were painful and time-consuming. With the TriVersa NanoMate®, we can run LC/fraction collection or infusion without changing the setup and wasting time with stabilization. We do not experience the problems typical with traditional nanoelectrospray sources.
One aspect of the TriVersa NanoMate® that impressed us was the ability to analyze complicated top-down samples with the LC compatibility. It is not possible to analyze these samples on an LC time scale, and the fraction collection capability allowed us to analyze in a way that was not possible previously.
To whom would you recommend the TriVersa NanoMate® for their research?
We use the TriVersa NanoMate® for everything; noncovalent interactions, top-down, middle-down, bottom-up, basic infusion, LC coupled to fraction collection. The instrument is useful in all of its different set-ups, especially without having to change sources and waiting for a stable spray.
The reliability of the system is one of the greatest benefits especially for people who have to change frequently between applications. We have found the spray sensing feature to be very valuable because we know our precious samples will not be lost.
Do you have any publications or presentations using the TriVersa NanoMate®?
Publication Highlight
Characterization of Human Sperm Protamine Proteoforms Through a Combination of Top-Down and Bottom-Up Mass Spectrometry Approaches
Soler-Ventura et al. J Proteome Res, 2020, 19(1), 221-237. DOI: 10.1021/acs.jproteome.9b00499
Identified the sperm protamine proteoforms profile, including their post-translational modifications, in normozoospermic individuals using a top-down MS approach and a proteinase-K-digestion-based bottom-up MS approach.
Other Publications
- Arauz-Garofalo et al. Protamine characterization by top-down proteomics: Boosting proteoform identification with DBSCAN. Proteoms, 2021. DOI: 10.3390/proteomes9020021
- Yero et al. The Pseudomonas aeruginosa substrate-binding protein Ttg2D functions as a general glycerophospholipid transporter across the periplasm. Comm Bio, 2021. DOI: 10.1038/s42003-021-01968-8
- Molnar et al. The histone code reader PHD finger protein 7 controls sex-linked disparities in gene expression and malignancy in Drosophila. Sci Adv, 2019. DOI: 10.1126/sciadv.aaw7965
- Nadal et al. Structure of the homodimeric androgen receptor ligand-binding domain. Nat Commun, 2017. DOI: 10.1038/ncomms14388
- Testoni et al. Lack of glycogenin causes glycogen accumulation and muscle function impairment. Cell Metabolism, 2017. DOI: 10.1016/j.cmet.2017.06.008
- Izquierdo-Serra et al. Optical control of endogenous receptors and cellular excitability using targeted covalent photoswitches. Nat Commun, 2016. DOI: 10.1038/ncomms12221
- Pujol-Pina et al. SDS-PAGE analysis of Aβ oligomers is disserving research into Alzheimer’s disease: Appealing for ESI-IM-MS. Sci Rep, 2015. DOI: 10.1038/srep14809
- Saez et al. Influence of PPh3 moiety in the anticancer activity of new organometallic ruthenium complexes. J Inorg Biochem, 2014. DOI: j.jinorgbio.2014.03.002
- Borg et al. Spectral counting assessment of protein dynamic range in cerebrospinal fluid following depletion with plasma-designed immunoaffinity columns. Clin Proteomics, 2011. DOI: 10.1186/1559-0275-8-6




