University of Cambridge, NIHR BCR Metabolomics and Lipidomics Facility
What is the focus of your lab’s research?
Almost exclusively high throughput lipid profiling of samples from large epidemiological studies, where we study gene-lifestyle interactions. The method works with plasma samples as well as dried blood spots. The method is also applied to small-scale studies of specific disease groups, dietary interventions and model systems such as yeast. The TriVersa NanoMate® is also used to study the lipid composition of tissues analyzed by LESA®.
Why did you incorporate the TriVersa NanoMate® into your laboratory?
The TriVersa NanoMate® is essential to efficiently analyze large-scale studies. It offers a really robust method for high throughput studies with minimal carryover.
Who would you recommend to purchase the TriVersa NanoMate®?
I recommend the TriVersa NanoMate® to laboratories with a large number of samples requiring a robust and reliable delivery system. The TriVersa NanoMate® eliminates typical nanoelectrospray ionization challenges.
Do you have any publications or presentations using the TriVersa NanoMate®?
Publication Highlight:
Development and Application of High-Throughput Single Cell Lipid Profiling: A Study of SNCA-A53T Human Dopamine Neurons
Snowden, et al. iScience, 2020, 23(10), 101703
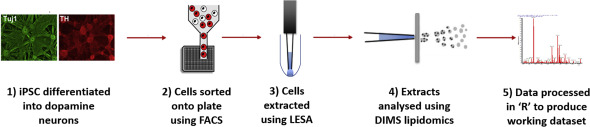
Combining FACS and LESA-MS to establish high-throughput single cell lipid profiling. Research identifies lipid differences found within and between populations of human dopamine neurons.
Other Publications:
- Snowden et al. Combining lipidomics and machine learning to measure clinical lipids in dried blood spots. Metabolomics. DOI: 10.1007/s11306-020-01703-0
- Koulman et al. The development and validation of a fast and robust dried blood spot based lipid profiling method to study infant metabolism. Metabolomics. DOI: 10.1007/s11306-014-0628-z
- Furse et al. A high-throughput platform for detailed lipidomic analysis of a range of mouse and human tissues. Analytical and Bioanalytical Chemistry. DOI: 10.1007/s00216-020-02511-0
- Harshfield et al. An unbiased lipid phenotyping approach to study the genetic determinants of lipids and their associations with coronary heart disease risk factors. J Proteom Res. DOI: 10.1021/acs.jproteome.8b00786
- Mann et al. Insights into genetic variants associated with NASH-fibrosis from metabolite profiling. Hum Mol Genet. DOI: 10.1093/hmg/ddaa162

